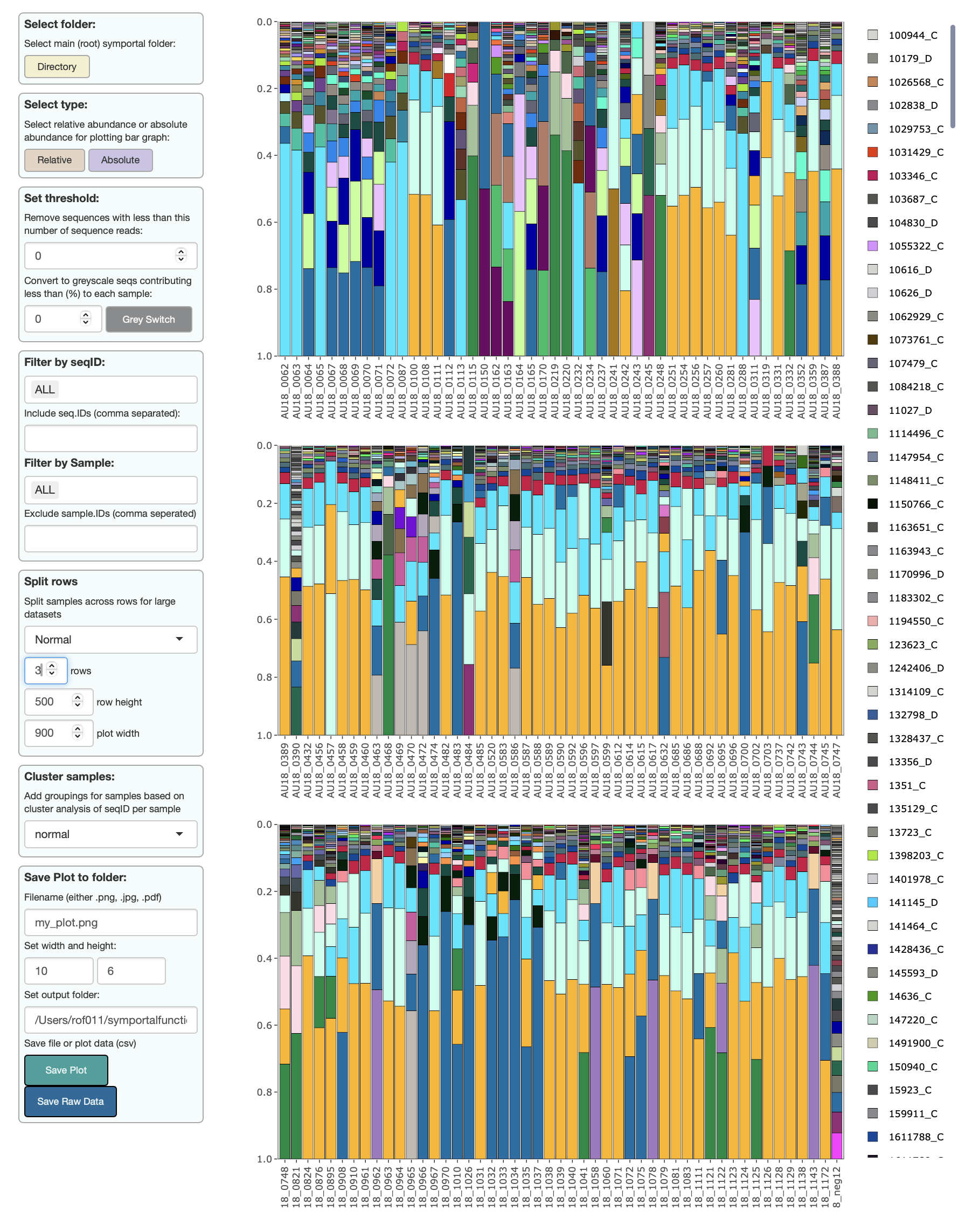
symbioR visualise sequences
symbioR includes a shiny app that can better visualise the ITS2 Symbiodinium community outputs from symportal. The shiny viewer allows:
uploading sequences via “select folder” for simple use
pplotlyto interact with plots to reveal rare seqID typesvisualisation of
relativevsabsolutesequence abundancessetting thresholds to remove rare sequences
setting thresholds to “ghost” rare sequences and enhance visualisation
filtering of plots by
seqIDfiltering of plots by
sample_namesplitting large datasets across multiple rows
ordering samples by hierarchical clustering of sequences (
bray-curtis,jaccard,euclidian,hellinger)saving plots to
png,jpg,pdfformats
library(tidyverse)
library(symbioR)
set.seed(1)
folder_path <- ("/symbiodinium/20230120T102936_esampayo")
view_sequences(folder_path)Or call view_sequences() with an empty plot and upload the folder using the folder window:
library(tidyverse)
library(symbioR)
set.seed(1)
folder_path <- ("/symbiodinium/20230120T102936_esampayo")
view_sequences(folder_path)