library(dplyr)
library(tidyr)
library(ggplot2)
library(stringr)
library(RColorBrewer)
symbio_pie <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") |>
select(sym_type_1, sym_type_2, sym_type_3) %>%
pivot_longer(cols = everything(), names_to = "variable", values_to = "combined") %>%
#mutate(combined = if_else(str_length(combined) <= 6, combined, NA_character_)) %>%
filter(!is.na(combined)) |>
filter(!grepl("rerun|mix\\?|_unidentified_band|Stereo", combined)) |>
group_by(combined) |>
summarise(count=n()) |>
ggplot() + theme_void() +
#ggtitle("Resolved sequences") +
ylab("") + xlab("") +
geom_bar(aes(x="", y=count, fill=combined), linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
scale_fill_manual(values = colorRampPalette(RColorBrewer::brewer.pal(12, "Paired"))(112)) +
theme(plot.title = element_text(size=12), legend.text = element_text(size=8), legend.key.size = unit(0.35, "cm"), legend.title=element_blank())
symbio_chart <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") %>%
select(sym_type_1, sym_type_2, sym_type_3) %>%
pivot_longer(cols = everything(), names_to = "variable", values_to = "combined") %>%
mutate(combined = if_else(str_length(combined) <= 6, combined, NA_character_)) %>%
filter(!is.na(combined)) %>%
group_by(combined) %>%
summarise(count = n()) %>%
arrange(desc(count)) %>%
slice_head(n = 10) %>%
arrange(combined) %>%
ggplot() + theme_bw() +
ggtitle("Top 10 most abundant sequence ID") +
ylab(" ") + xlab("") +
geom_bar(aes(x = combined, y = count, fill = combined), linewidth = 0.1, show.legend=FALSE,
color = "black", stat = "identity", width = 1) +
scale_fill_manual(values = colorRampPalette(RColorBrewer::brewer.pal(12, "Paired"))(10)) +
theme(
plot.title = element_text(size = 4),
legend.text = element_text(size = 4),
legend.title = element_blank(),
axis.text.x = element_text(angle = 45, hjust = 1)
)
symbio_pie + plot_annotation(title='Symbiobase collection of Symbiodiniaceae by sequence ID') &
theme(plot.title = element_text(size = 11))

library(tidyverse)
library(lubridate)
library(ggplot2)
library(plotly)
library(patchwork)
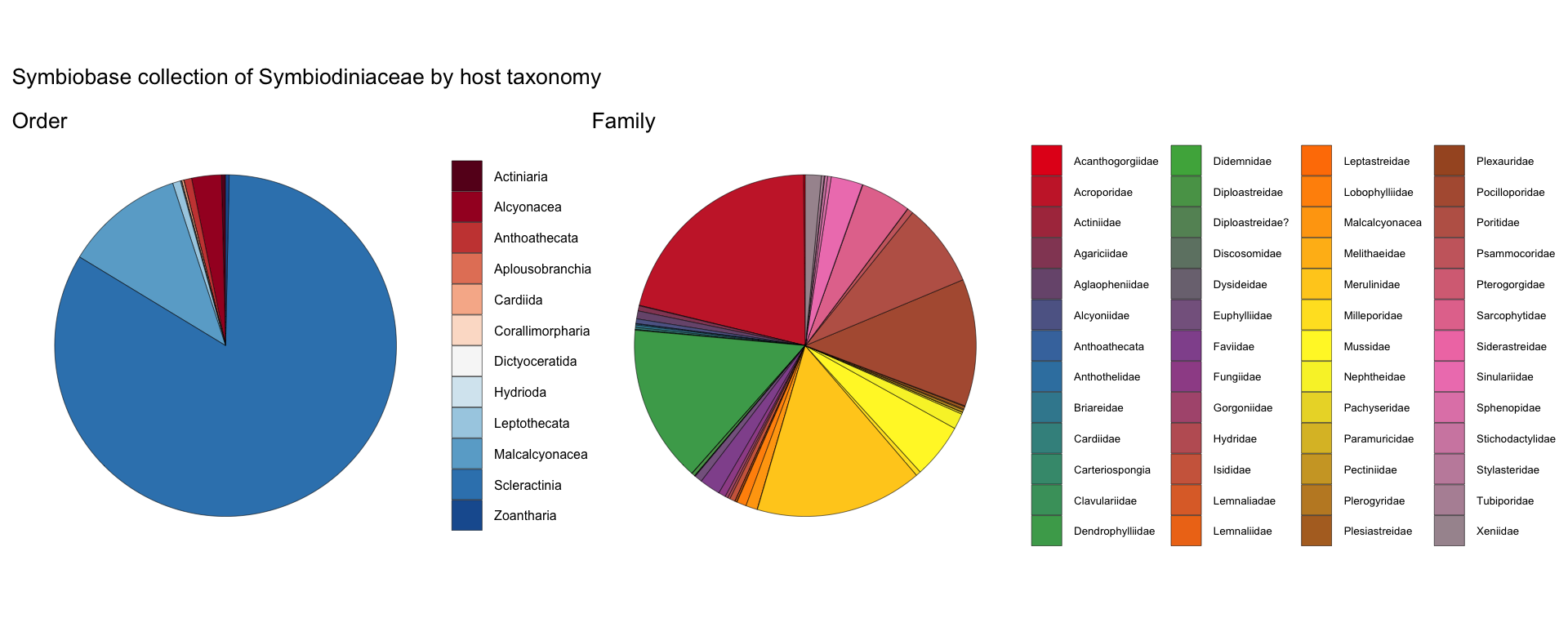
order <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") |>
dplyr::select(host_order) |>
drop_na(host_order) |>
group_by(host_order) |>
summarise(count=n()) |>
ggplot() + theme_void() +
ggtitle("Order") + ylab("") + xlab("") +
geom_bar(aes(x="", y=count, fill=host_order), linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
#geom_text(data=data_pie_latitude, aes(x="", y=count, label = count), size=6, position = position_stack(vjust = 0.5)) +
scale_fill_manual(values = colorRampPalette(RColorBrewer::brewer.pal(13, "RdBu"))(13)) +
theme(plot.title = element_text(size=6), legend.text = element_text(size=6), legend.key.size = unit(0.5, "cm"), legend.title=element_blank())
family <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") |>
dplyr::select(host_family) |>
drop_na(host_family) |>
group_by(host_family) |>
summarise(count=n()) |>
ggplot() + theme_void() +
ggtitle("Family") + ylab("") + xlab("") +
geom_bar(aes(x="", y=count, fill=host_family), linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
scale_fill_manual(values = colorRampPalette(RColorBrewer::brewer.pal(12, "Set1"))(53)) +
theme(plot.title = element_text(size=6), legend.text = element_text(size=5), legend.title=element_blank(), legend.key.size = unit(0.5, "cm") ) +
guides(fill = guide_legend(ncol = 4))
order + family + plot_annotation(title='Symbiobase collection of Symbiodiniaceae by host taxonomy') &
theme(plot.title = element_text(size = 10))