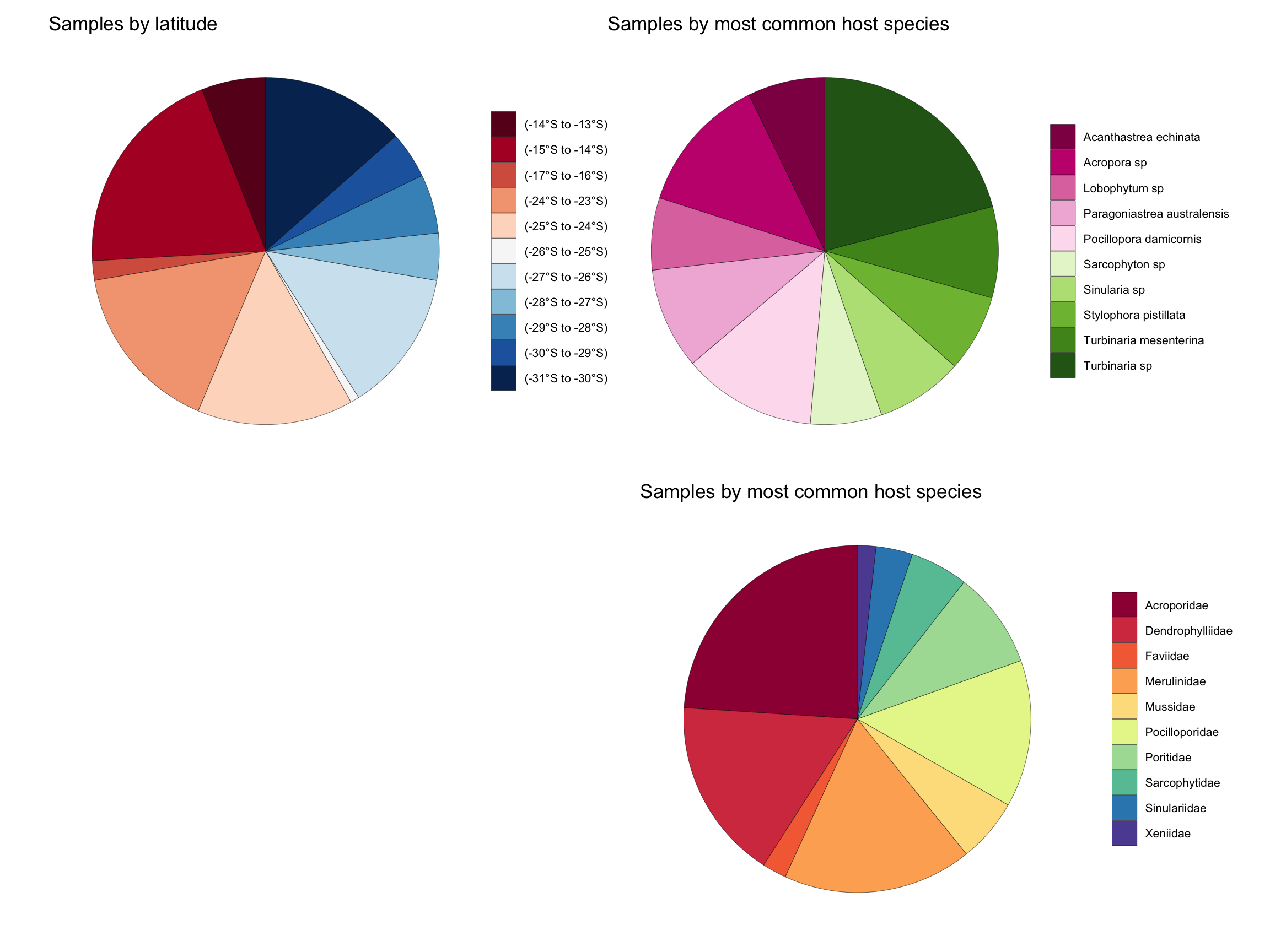
Collection statistics
The collection by numbers (01/01/2024):
4143 samples across the Eastern Australian coastline
Subtropical to tropical collections, spanning 20° of latitude (-11.4°S to -30.94°S)
54 host families
279 host species
library(tidyverse)
library(lubridate)
library(ggplot2)
library(plotly)
data_pie_latitude <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") |>
dplyr::select(sample_id, collection_date, location, site, longitude, latitude, host_family, host_genus, host_species) |>
mutate(latitude=round(latitude,3), longitude=round(longitude,3), collection_date=ymd(collection_date)) |>
mutate(location = gsub("_", " ", location),
site = gsub("_", " ", site),
host_genus = gsub("_", " ", host_genus),
host_species = gsub("_", " ", host_species)) |>
# drop_na(lat) %>%
mutate(latitude_groups = cut(latitude, breaks = seq(-34, -13, by = 1), include.lowest = TRUE)) %>%
filter(!latitude_groups=="NA") |>
mutate(latitude_groups = gsub(",", " to ", latitude_groups) %>%
gsub("\\]", ")", .) %>%
gsub("(-?\\d+)", "\\1°S", .)) |>
group_by(latitude_groups) |>
summarise(count=n())
plot_lat <- ggplot() + theme_void() +
ggtitle("Samples by latitude") + ylab("") + xlab("") +
geom_bar(data=data_pie_latitude, aes(x="", y=count, fill=latitude_groups), linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
#geom_text(data=data_pie_latitude, aes(x="", y=count, label = count), size=6, position = position_stack(vjust = 0.5)) +
scale_fill_brewer(palette="RdBu") + theme(legend.text = element_text(size=8), legend.title=element_blank())
data_pie_species <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") %>%
dplyr::select(sample_id, collection_date, location, site, longitude, latitude, host_family, host_genus, host_species) %>%
mutate(
latitude = round(latitude, 3),
longitude = round(longitude, 3),
collection_date = ymd(collection_date),
location = gsub("_", " ", location),
site = gsub("_", " ", site),
host_genus = gsub("_", " ", host_genus),
host_species = gsub("_", " ", host_species),
latitudinal_band = as.character(cut(latitude, breaks = seq(-34, -13, by = 1), include.lowest = TRUE))
) %>%
mutate(latitudinal_band = gsub(",", " to ", latitudinal_band) %>%
gsub("\\]", ")", .) %>%
gsub("(-?\\d+)", "\\1°S", .)) %>%
filter(!host_species %in% c("sp1", "sp2")) %>%
mutate(host_species = factor(host_species, levels = sort(unique(host_species), decreasing = FALSE))) %>%
drop_na(host_species) %>%
group_by(host_species) %>%
summarise(count = n()) %>%
slice_max(order_by = count, n = 10)
plot_species <- ggplot() + theme_void() +
ggtitle("Samples by most common host species") + ylab("") + xlab("") +
geom_bar(data=data_pie_species, aes(x="", y=count, fill=host_species),
show.legend=TRUE, linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
scale_fill_brewer(palette="PiYG") + theme(legend.text = element_text(size=8), legend.title=element_blank())
data_pie_family <- read.csv("https://raw.githubusercontent.com/symbiobase/symbioID/main/Symbiobase.csv") |>
dplyr::select(sample_id, collection_date, location, site, longitude, latitude, host_family, host_genus, host_species) |>
mutate(latitude=round(latitude,3), longitude=round(longitude,3), collection_date=ymd(collection_date)) |>
mutate(location = gsub("_", " ", location),
site = gsub("_", " ", site),
host_genus = gsub("_", " ", host_genus),
host_species = gsub("_", " ", host_species)) |>
mutate(latitudinal_band = as.character(cut(latitude, breaks = seq(-34, -13, by = 1), include.lowest = TRUE))) %>%
mutate(latitudinal_band = gsub(",", " to ", latitudinal_band) %>%
gsub("\\]", ")", .) %>%
gsub("(-?\\d+)", "\\1°S", .)) %>%
filter(!host_species %in% c("sp1", "sp2")) %>%
mutate(host_family = factor(host_family, levels = sort(unique(host_family), decreasing = FALSE))) |>
drop_na(host_family) |>
group_by(host_family) %>%
summarise(count = n()) %>%
slice_max(order_by = count, n = 10)
plot_family <- ggplot() + theme_void() +
ggtitle("Samples by most common host species") + ylab("") + xlab("") +
geom_bar(data=data_pie_family, aes(x="", y=count, fill=host_family),
show.legend=TRUE, linewidth=0.1, color="black", stat="identity", width=1) +
coord_polar("y", start=0) +
scale_fill_brewer(palette="Spectral") + theme(legend.text = element_text(size=8), legend.title=element_blank())
library(patchwork)
(plot_lat + plot_species) / (plot_spacer()+plot_family)